Performs pixelwise updates based on conditional distributions to sample from a Markov random field.
rmrf2d(
init_Z,
mrfi,
theta,
cycles = 60,
sub_region = NULL,
fixed_region = NULL
)Arguments
- init_Z
One of two options:
A
matrixobject with the initial field configuration. Its valuesmust be integers in{0,...,C}.A length 2
numericvector with the lattice dimensions.
- mrfi
A
mrfiobject representing the interaction structure.- theta
A 3-dimensional array describing potentials. Slices represent interacting positions, rows represent pixel values and columns represent neighbor values. As an example:
theta[1,3,2]has the potential for the pair of values 0,2 observed in the second relative position ofmrfi.- cycles
The number of updates to be done (for each each pixel).
- sub_region
NULLif the whole lattice is considered or alogicalmatrixwithTRUEfor pixels in the considered region.- fixed_region
NULLif the whole lattice is to be sampled or alogicalmatrixwithTRUEfor pixels to be considered fixed. Fixed pixels are not updated in the Gibbs Sampler.
Value
A matrix with the sampled field.
Details
This function implements a Gibbs Sampling scheme to sample from a Markov random field by iteratively sampling pixel values from the conditional distribution $$P(Z_i | Z_{{N}_i}, \theta).$$
A cycle means exactly one update to each pixel. The order pixels are sampled is randomized within each cycle.
If init_Z is passed as a length 2 vector with lattice dimensions, the
initial field is sampled from independent discrete uniform distributions in
{0,...,C}. The value of C is obtained from the number of rows/columns of
theta.
A MRF can be sampled in a non-rectangular region of the lattice with the use of
the sub_region argument or by setting pixels to NA in the initial
configuration init_Z. Pixels with NA values in init_Z are completely
disconsidered from the conditional probabilities and have the same effect as
setting sub_region = is.na(init_Z). If init_Z has NA values,
sub_region is ignored and a warning is produced.
A specific region can be kept constant during the Gibbs Sampler by using the
fixed_region argument. Keeping a subset of pixels constant is useful when
you want to sample in a specific region of the image conditional to the
rest, for example, in texture synthesis problems.
Note
As in any Gibbs Sampling scheme, a large number of cycles may be required to achieve the target distribution, specially for strong interaction systems.
See also
A paper with detailed description of the package can be found at doi: 10.18637/jss.v101.i08 .
rmrf2d_mc for generating multiple points of a
Markov Chain to be used in Monte-Carlo methods.
Examples
# Sample using specified lattice dimension
Z <- rmrf2d(c(150,150), mrfi(1), theta_potts)
#Sample using itial configuration
# \donttest{
Z2 <- rmrf2d(Z, mrfi(1), theta_potts)
# View results
dplot(Z)
 dplot(Z2)
dplot(Z2)
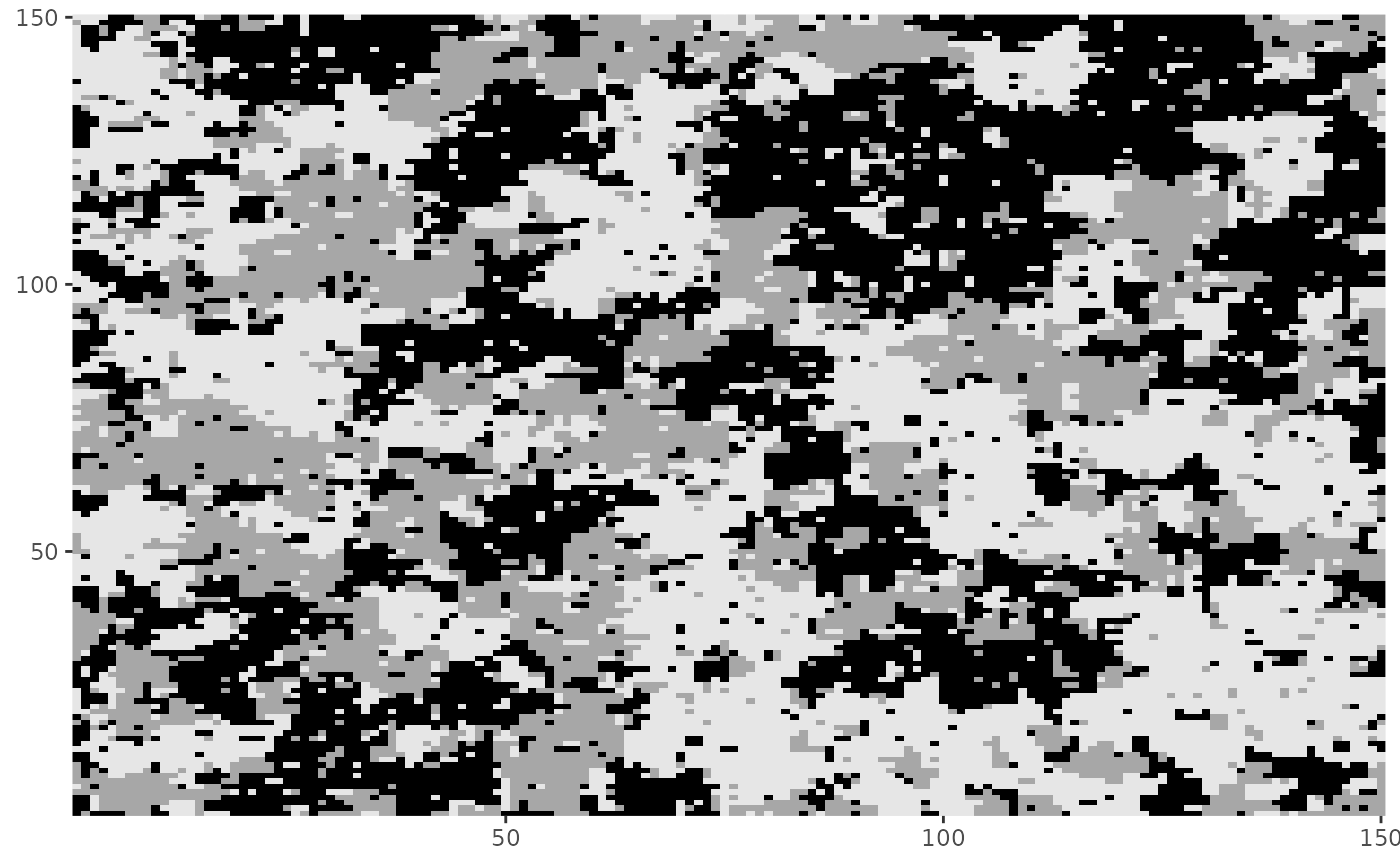 # Using sub-regions
subreg <- matrix(TRUE, 150, 150)
subreg <- abs(row(subreg) - 75) + abs(col(subreg) - 75) <= 80
# view the sub-region
dplot(subreg)
# Using sub-regions
subreg <- matrix(TRUE, 150, 150)
subreg <- abs(row(subreg) - 75) + abs(col(subreg) - 75) <= 80
# view the sub-region
dplot(subreg)
 Z3 <- rmrf2d(c(150,150), mrfi(1), theta_potts, sub_region = subreg)
dplot(Z3)
Z3 <- rmrf2d(c(150,150), mrfi(1), theta_potts, sub_region = subreg)
dplot(Z3)
 # Using fixed regions
fixreg <- matrix(as.logical(diag(150)), 150, 150)
# Set initial configuration: diagonal values are 0.
init_Z4 <- Z
init_Z4[fixreg] <- 0
Z4 <- rmrf2d(init_Z4, mrfi(1), theta_potts, fixed_region = fixreg)
dplot(Z4)
# Using fixed regions
fixreg <- matrix(as.logical(diag(150)), 150, 150)
# Set initial configuration: diagonal values are 0.
init_Z4 <- Z
init_Z4[fixreg] <- 0
Z4 <- rmrf2d(init_Z4, mrfi(1), theta_potts, fixed_region = fixreg)
dplot(Z4)
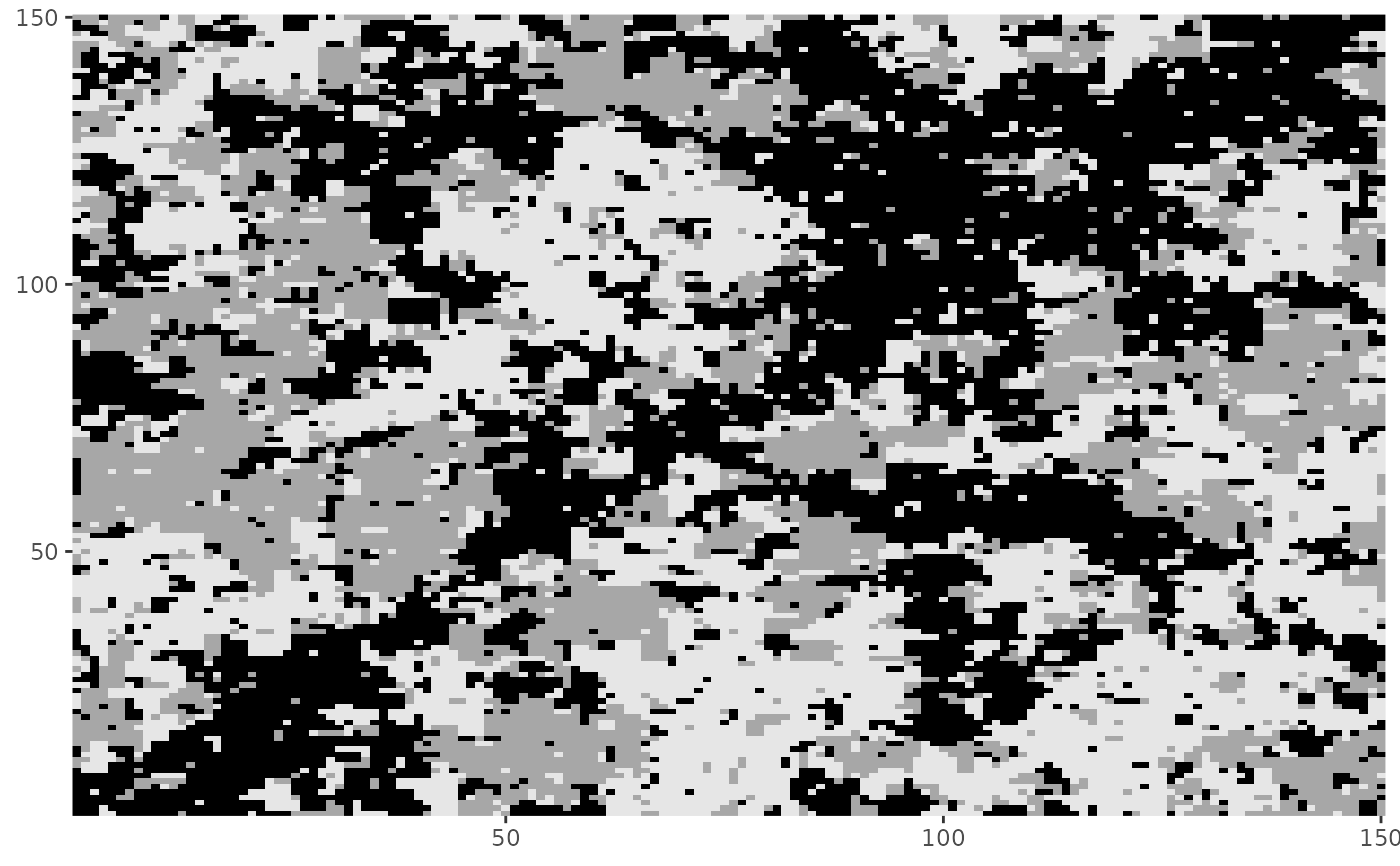 # Combine fixed regions and sub-regions
Z5 <- rmrf2d(init_Z4, mrfi(1), theta_potts,
fixed_region = fixreg, sub_region = subreg)
#> Warning: Some pixels in the 'fixed_region' are not part of the 'sub_region', they will be ignored.
dplot(Z5)
# Combine fixed regions and sub-regions
Z5 <- rmrf2d(init_Z4, mrfi(1), theta_potts,
fixed_region = fixreg, sub_region = subreg)
#> Warning: Some pixels in the 'fixed_region' are not part of the 'sub_region', they will be ignored.
dplot(Z5)
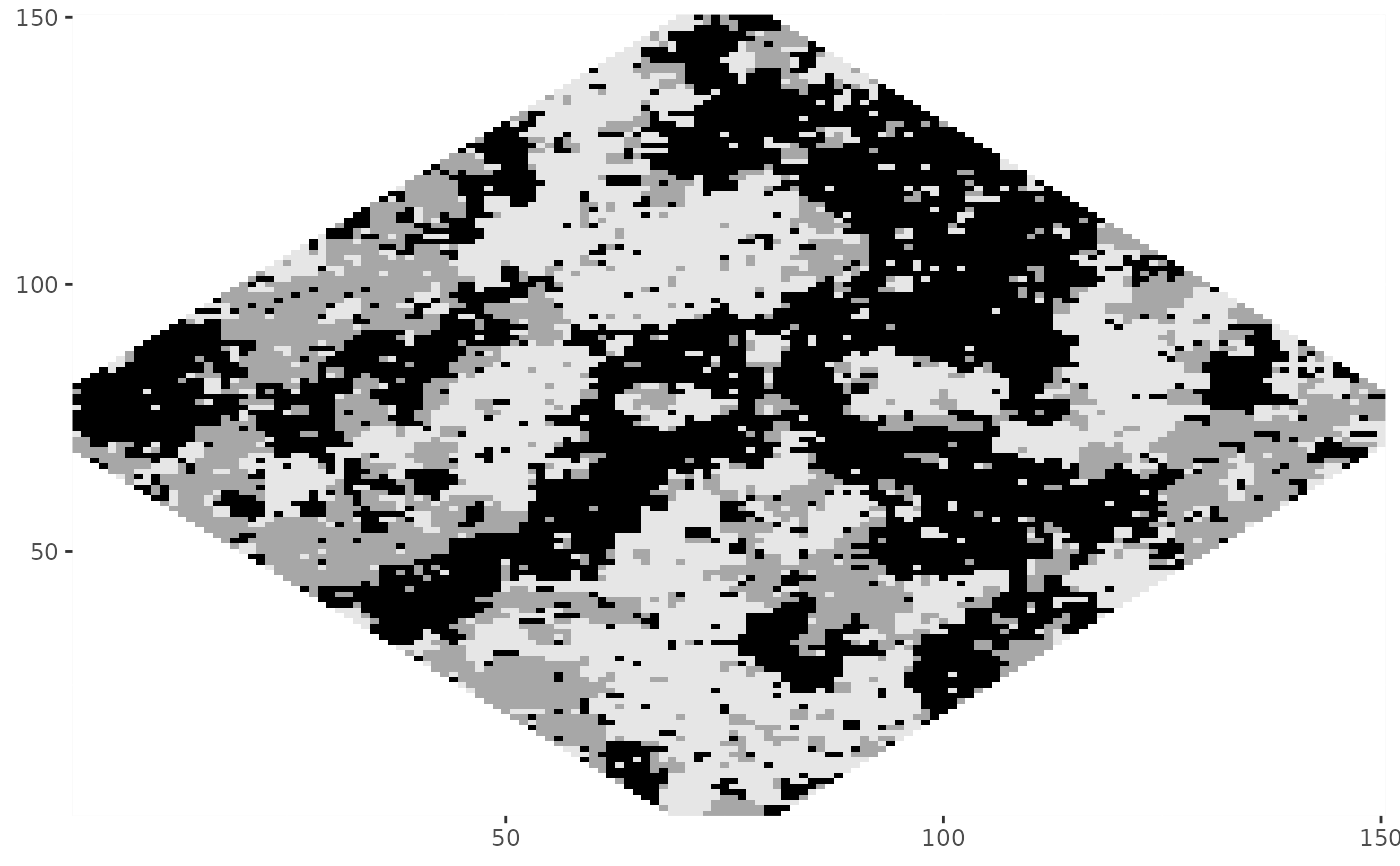 # }
# }