fit_ghm fits a Gaussian Mixture model with hidden components
driven by a Markov random field with known parameters. The inclusion of a
linear combination of basis functions as a fixed effect is also possible.
The algorithm is a modification of of (Zhang et al. 2001) , which is described in (Freguglia et al. 2020) .
fit_ghm(
Y,
mrfi,
theta,
fixed_fn = list(),
equal_vars = FALSE,
init_mus = NULL,
init_sigmas = NULL,
maxiter = 100,
max_dist = 10^-3,
icm_cycles = 6,
verbose = interactive(),
qr = NULL
)Arguments
- Y
A matrix of observed (continuous) pixel values.
- mrfi
A
mrfiobject representing the interaction structure.- theta
A 3-dimensional array describing potentials. Slices represent interacting positions, rows represent pixel values and columns represent neighbor values. As an example:
theta[1,3,2]has the potential for the pair of values 0,2 observed in the second relative position ofmrfi.- fixed_fn
A list of functions
fn(x,y)to be considered as a fixed effect. Seebasis_functions.- equal_vars
logicalindicating if the mixture model has the same variances in all mixture components.- init_mus
Optional. A
numericwith length (C+1) with the initial mean estimate for each component.- init_sigmas
Otional. A
numericwith length (C+1) with initial sample deviation estimate for each component.- maxiter
The maximum number of iterations allowed. Defaults to 100.
- max_dist
Defines a stopping condition. The algorithm stops if the maximum absolute difference between parameters of two consecutive iterations is less than
max_dist.- icm_cycles
Number of steps used in the Iterated Conditional Modes algorithm executed in each interaction. Defaults to 6.
- verbose
logicalindicating wheter to print the progress or not.- qr
The QR decomposition of the design matrix. Used internally.
Value
A hmrfout containing:
par: Adata.framewith \(\mu\) and \(\sigma\) estimates for each component.fixed: Amatrixwith the estimated fixed effect in each pixel.Z_pred: Amatrixwith the predicted component (highest probability) in each pixel.predicted: Amatrixwith the fixed effect + the \(\mu\) value for the predicted component in each pixel.iterations: Number of EM iterations done.
Details
If either init_mus or init_sigmas is NULL an EM algorithm
considering an independent uniform distriburion for the hidden component is
fitted first and its estimated means and sample deviations are used as
initial values. This is necessary because the algorithm may not converge if
the initial parameter configuration is too far from the maximum likelihood
estimators.
max_dist defines a stopping condition. The algorithm will stop if the
maximum absolute difference between (\(\mu\) and \(\sigma\)) parameters
in consecutive iterations is less than max_dist.
References
Freguglia V, Garcia NL, Bicas JL (2020).
“Hidden Markov random field models applied to color homogeneity evaluation in dyed textile images.”
Environmetrics, 31(4), e2613.
Zhang Y, Brady M, Smith S (2001).
“Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm.”
IEEE transactions on medical imaging, 20(1), 45--57.
See also
A paper with detailed description of the package can be found at doi: 10.18637/jss.v101.i08 .
Examples
# Sample a Gaussian mixture with components given by Z_potts
# mean values are 0, 1 and 2 and a linear effect on the x-axis.
# \donttest{
set.seed(2)
Y <- Z_potts + rnorm(length(Z_potts), sd = 0.4) +
(row(Z_potts) - mean(row(Z_potts)))*0.01
# Check what the data looks like
cplot(Y)
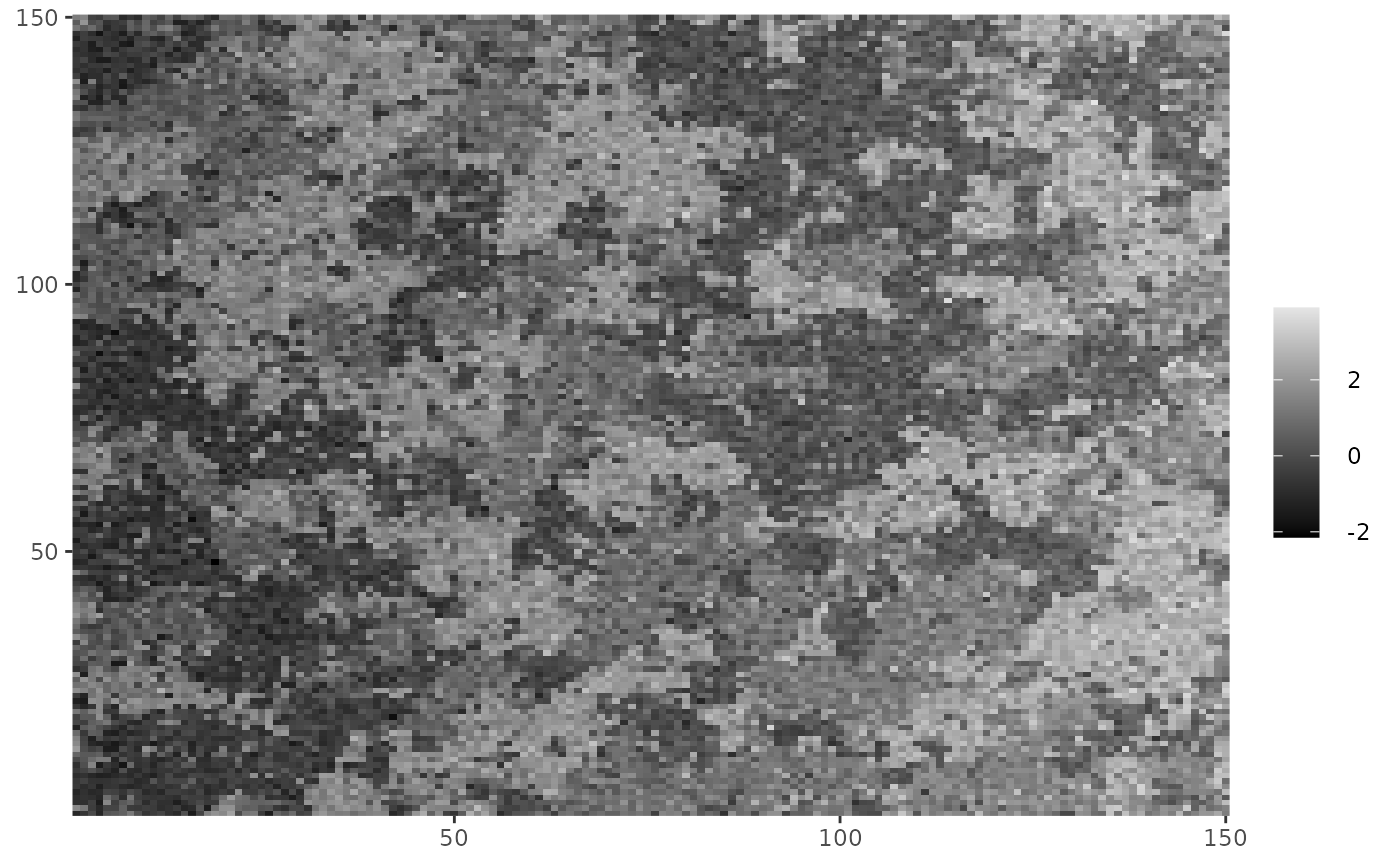 fixed <- polynomial_2d(c(1,0), dim(Y))
fit <- fit_ghm(Y, mrfi = mrfi(1), theta = theta_potts, fixed_fn = fixed)
#>
Fitting independent mixture to obtain initial parameters.
#>
EM iteration: 1
EM iteration: 2
EM iteration: 3
EM iteration: 4
EM iteration: 5
EM iteration: 6
Finished with 6 iterations.
fit$par
#> mu sigma
#> 0 -0.001568028 0.4012135
#> 1 0.999990536 0.3966913
#> 2 2.002972466 0.4026593
cplot(fit$fixed)
fixed <- polynomial_2d(c(1,0), dim(Y))
fit <- fit_ghm(Y, mrfi = mrfi(1), theta = theta_potts, fixed_fn = fixed)
#>
Fitting independent mixture to obtain initial parameters.
#>
EM iteration: 1
EM iteration: 2
EM iteration: 3
EM iteration: 4
EM iteration: 5
EM iteration: 6
Finished with 6 iterations.
fit$par
#> mu sigma
#> 0 -0.001568028 0.4012135
#> 1 0.999990536 0.3966913
#> 2 2.002972466 0.4026593
cplot(fit$fixed)
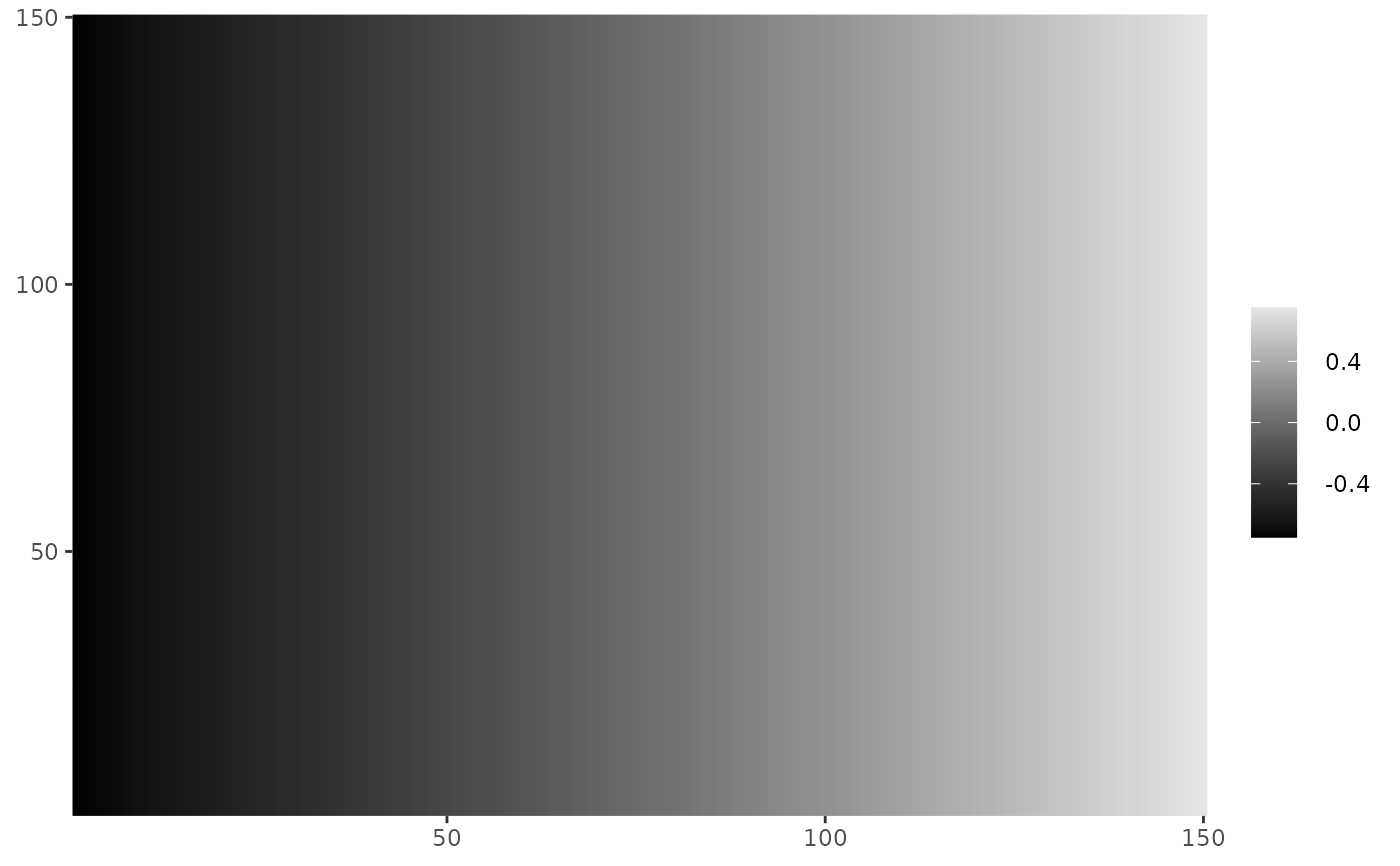 # }
# }